|
|
Biology of Methanogenesis |
Department of Marine Biotechnology UMBC - Institute of Marine & Environmental Technology
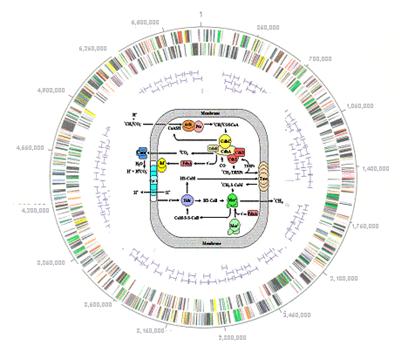
Global regulatory pathways for methanogenesis
The broad objective is an academic-industry collaboration with investigators at Penn State University, University of California at Los Angeles and Dupont to exploit the genomic sequence of Methanosarcina thermophila with the immediate goal of discovering global principals and novel genes/proteins for two key physiological processes, and also to provide a foundation for the longer-term goal of discovering global principals and novel molecular biology that regulate these processes. A functional genomics approach will be taken to (i) further our understanding of the fermentation of acetate to methane, the pathway by which two-thirds of the global output of methane is produced, and (ii) how M. thermophila (and the Archaea in general) respond to environmental stress. By understanding these physiological processes in greater detail, the results are expected to increase our understanding of biocomplexity by learning how M. thermophila reacts to changing environmental conditions and how it interacts with other members in microbial consortia that comprise the anaerobic link in the global carbon cycle.
The specific aims are 1) employ DNA microarray
and proteomics identify genes specific for methane formation from acetate
and uncover general principals of how Archaea adapt to changing environmental
conditions; 2) conduct transcriptional mapping and comparative gene sequence
analysis to determine potential gene function and lay a foundation for the
longer-term goal of understanding the molecular mechanisms of regulation in
the Archaea domain; 3) conduct gene knockout experiments to confirm the specific
function of selected genes; 4) over-express gene products for biochemical
and structural analysis. A large percentage of the open reading frames of
sequenced microbial genomes, including M. thermophila, have no significant
deduced identity to any known proteins; thus it is expected that novel proteins
and enzymes will be discovered and that their characterization will uncover
new biochemical principals.
 Collaborators
Collaborators
James G. Ferry, Ph.D. – Penn State University
Robert P. Gunsalus – University of California at Los Angeles
Project Team
Kimberly Anderson, B.S., M.S.
Sheridan MacAuley, B.S.
Publications and Presentations
MacAuley, S.R., S.A. Zimmerman, E.E. Apolinario, C. Evilia, Y.-M. Hou, J.G. Ferry, K.R. Sowers. The Archetype g-Class Carbonic Anhydrase (Cam) Contains Iron when Synthesized in vivo in Methanosarcina acetivorans. Biochemistry 48(5): 817-9 [ABSTRACT].
Sowers, K.R. 2009. Methanogenesis. In: Schaechter, Lederberg, Alexander, Haselkorn, Ingraham, Baross, Schmidt, Laskin, Hopwood, Summers, Baulcombe, Levin, White, Fierer, Baldauf (eds.), Encylopedia of Microbiology, 3rd edition. Elsevier, Inc. ISBN: 978-0-12-373944-5.
Sowers, K.R. and K. Anderson. 2007. Molecular Genetics of Archaea. In: R. Cavicchioli (ed.), Archaea: Molecular Cell Biology. American Society for Microbiology, Washington, D.C. , pp. 463-477. ISBN: 978-1-55581-391-8.
Sowers, K.R., S. DasSarma and P. Blum. 2007. Gene transfer in Archaea. In: C. A. Reddy, T. J. Beveridge, J. A. Breznak, G. A. Marzluf, and T. M. Schmidt (ed.), Methods for General and Molecular Microbiology. American Society for Microbiology, Washington, D. C. ISBN: 978-1-55581-223-2.
Maeder, D.L., I. Anderson, T. Brettin, D. Bruce, P. Gilna, C. S. Han, A. Lapidus, W.W. Metcalf, E. Saunders, R. Tapia, and K.R. Sowers. 2006. The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J. Bacteriol. 188: 7922-7931.
Related Publications and Abstracts
Li, L., Li, Q., Rohlin, L., Kim, U., Salmon, K., Rejtar, T., Gunsalus, R.P., Karger, B.L., Ferry, J.G. 2007. Quantitative Proteomic and Microarray Analysis of the Archaeon Methanosarcina acetivorans Grown with Acetate versus Methanol. J. Proteome Res. 6: 759-771.
Li, Q., Li, Lngyun, Rejtar, T., Karger, B.L., Ferry, J.G. Proteome of Methanosarcina acetivorans Part I: An Expanded View of the Biology of the Cell. J. Proteome Res. 4: 112-128.
Li, Q., Li, Lngyun, Rejtar, T., Karger, B.L., Ferry, J.G. Proteome of Methanosarcina acetivorans Part II: Comparison of Protein Levels in Acetate- and Methanol-Grown Cells. J. Proteome Res. 4: 129-135.
Galagan, J.E., et al. 2002. The genome of Methanosarcina acetivorans reveals extensive metabolic and physiological diversity. Genome Research 12: 532-542.
Jackson,
K., Apolinario-Smith, E. and K.R. Sowers.
2002.
Regulation
of Catabolic Carbon
Monoxide Dehydrogenase in the Archaeon Methanosarcina acetivorans.
Southeastern Branch of
the Amercian Society of Microbiology Annual Meeting, Gainsville,
Florida.
November 7-9.
Funded by