problem-spec.c into
individual problem specification files
square-spec.cannulus-spec.ctriangle-with-hole-spec.cthree-holes-spec.c
problem-spec.h intact because it is
domain-independent and defines the structures that are needed
in *-spec.c files.
This project focuses on the square and annulus domains for which we have exact solutions. Do the other two domains also if you wish, but that's optional.
Make a header file for each *-spec.c file.
Here is the complete annulus-spec.h:
#ifndef H_ANNULUS_SPEC_H #define H_ANNULUS_SPEC_H #include "problem-spec.h" struct problem_spec *get_spec_annulus(int n); void free_annulus(struct problem_spec *spec); #endif // H_ANNULUS_SPEC_H
-
On the $(0,1)\times(0,1)$ square take
$f(x,y) = 32 \bigl(x(1-x) + y(1-y)\bigr)$.
This corresponds to the exact solution
$u(x,y) = 16x(1-x)y(1-y)$ which you may use
to ascertain the solver's accuracy.
Thus, in
square-spec.cdefinestatic double f(double x, double y) { return 32*(x*(1-x) + y*(1-y)); } static double exact(double x, double y) { return 16*x*y*(1-x)*(1-y); }and then insertf()andexact()in theproblem_specstructure:static struct problem_spec spec = { .points = points, .segments = segments, .holes = NULL, .npoints = (sizeof points)/(sizeof points[0]), .nsegments = (sizeof segments)/(sizeof segments[0]), .nholes = 0, .f = f, // here it is! .g = NULL, .h = NULL, .eta = NULL, .u_exact = exact, // here it is! };Here is what I get:
./demo-square 5 0.01 domain is a square nodes = 96, edges = 254, elems = 159 system stiffness matrix is 96x96 (=9216) has 424 nonzero entries errors: L^infty = 0.0413703, L^2 = 0.0154083, energy norm = 0.388679 pyplot output written to file demo-square.py
Verify that the $L^2$ error is proportional to $a$ (the maximal triangle area) as $a$ approaches zero.
-
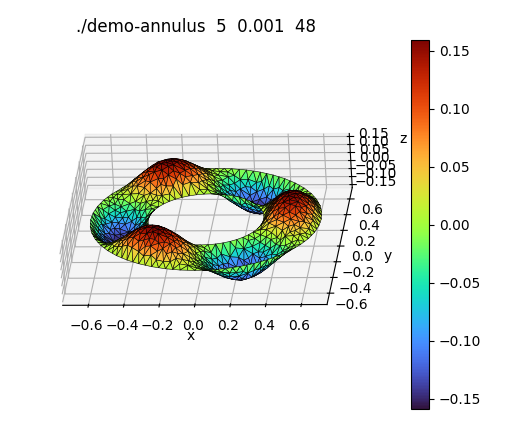
On the
annulus, take $\ds f(x,y) = \frac{s}{r^2} \Bigl[ 8 (a+b) r - 9 a b - 5 r^2 \Bigr] \cos3t$, where $a$ and $b$ are the inner and outer radii, and $s = 2/(b-a)$.Here $f$ is defined in terms of the polar coordinates $r$ and $t$. We have $r = \sqrt{x^2+y^2}$ and $t = \tan^{-1}(y/x)$. Use the C Standard Library's
atan2for the inverse tangent, as int = tan2(y,x).The exact solution in this case is $u=s(r-a)(b-r) \cos 3t$. Use that to determine the accuracy of the solver.
Due to poor design, our function $f$ does not take optional arguments as parameters. (Refer to the Nelder-Mead project to see how optional parameters are handled.) For now, you may set the parameters $a$, $b$, and $s$ as preprocessor macros in
annuluc-spec.c, as in#define a 0.325 #define b (2*a) #define s 2.0/(b-a)
That works, but such abuse of preprocessor macros is considered poor style. If you feel motivated, see if you can change things to allow parameters in each of the functionsf,g,h,η,u_exactthat are declared inproblem-spec.h. For instance, you may want to change the linedouble (*f)(double x, double y);
todouble (*f)(double x, double y, void *params);
to allow passing parameters tof.Here is what I get:
./demo-annulus 5 0.001 48 domain is an annulus (really a 48-gon) of radii 0.325 and 0.65 nodes = 860, edges = 2436, elems = 1576 system stiffness matrix is 860x860 (=739600) has 4830 nonzero entries errors: L^infty = 0.00554751, L^2 = 0.00199565, energy norm = 0.146381 pyplot output written to file demo-annulus.py
-
[Optional] On
triangle-with-holetake $f(x,y) = 30 x y (2-x) (2-y)$. -
[Optional] On
three-holestake $f(x,y) = 15.0$.
plot-with-python.c and plot-with-python.h
that provide a function plot_with_python() which receives
the solution of your FEM solver and writes a python script for plotting
the solution's 3D graph. Here is my plot-with-python.h:
#ifndef H_PLOT_WITH_PYTHON_H #define H_PLOT_WITH_PYTHON_H #include "mesh.h" void plot_with_python(struct mesh *mesh, char **argv, char *filename); #endif // H_PLOT_WITH_PYTHON_HThe purpose of passing
argv to that function is so that
it can put the command-line arguments in the graph's title (see below).
The filename argument is the name of the output file.
Here is an sample python script that plots a surface consisting of two triangles.
# --- A --------------------------------#
import numpy as np
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
from matplotlib import cm
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
ax.set_proj_type('ortho')
# --- B --------------------------------#
ax.set_title('Your Title Here') // optional: set title
coords = np.array([
[0, 0, 0],
[1, 0, 0],
[1, 1, 1],
[0, 1, 0.5],
])
elems = np.array([
[0, 1, 2],
[0, 2, 3],
])
# --- C --------------------------------#
x = coords[:, 0]
y = coords[:, 1]
z = coords[:, 2]
surf = ax.plot_trisurf(x, y, z, triangles=elems,
cmap=cm.turbo, edgecolor='black', linewidth=0.4)
fig.colorbar(surf, shrink=0.75)
ax.set_xlabel('x')
ax.set_ylabel('y')
ax.set_zlabel('z')
ax.set_aspect('equal') # requires matplotlib version >= 3.6
plt.show()
# --------------------------------------#
The group of lines marked A and C are independent of the
particular problem. Only those in group B vary.
The array coords consists of the coordinates
$[x_i, y_i, z_i]$, $i=0,1,2,\ldots$ of the surface's points.
The array elems consists of the indices
$[v^{(i)}_0, v^{(i)}_1, v^{(i)}_2]$, $i=0,1,2,\dots$ of element vertices.
Optionally, you may
set the plot's title through the set_title line
shown above.
I like to set it to command-line arguments which are available through
the argv array in main(). Here is a sample: