

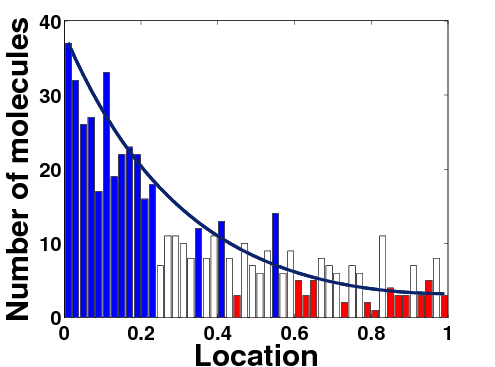
 (blue, white, and red: different cell types based on the morphogen molecule number) |
Boundary formation between different cell types involves spatially-distributed signals called morphogens that influence the phenotypic identity of cells. Our question is how sensitive the boundary location is to variations in parameter values in the system, such as reaction and diffusion rates, size of the system, and input flux. We analyze both deterministic and stochastic reaction-diffusion models to understand how the signaling scheme affects the variability of boundary determination between cell types. Our objective in the project was to understand how the placement of the morphogen sources and the type of interpretation function used as the response affect the location of specified level sets of the response. In the stochastic framework we developed a new method for defining the boundary between cell types by appropriately quantizing the probability of exceeding a specified threshold in the number of a downstream molecule. |